Homepage of Hannes Nickisch
 After doing a PhD at the Max Planck
Institute for Biological Cybernetics and a PostDoc at the Max
Planck Institue for Intelligent Systems, I'm using machine learning and AI methods for
medical image analysis tasks e.g. for cardiovascular1,2 and oncology applications as a Senior Scientist
at Philips, Hamburg, Germany.
After doing a PhD at the Max Planck
Institute for Biological Cybernetics and a PostDoc at the Max
Planck Institue for Intelligent Systems, I'm using machine learning and AI methods for
medical image analysis tasks e.g. for cardiovascular1,2 and oncology applications as a Senior Scientist
at Philips, Hamburg, Germany.
During my study times at the Technical University Berlin and the Université de Nantes I did internships with Microsoft Research in Cambridge, Siemens Corporate Research in Princeton and Siemens Medical Systems in Erlangen.
My research interests lie in probabilistic machine learning, computer vision and pattern recognition, medical image processing and predictive patient-specific biophysical models based on Gaussian processes and neural networks
My work is listed below, and on
![]()
![]()
![]()
![]()
![]() .
In case you wish to collaborate or have questions, please contact me via
contact.hannes@nickisch.org or
.
In case you wish to collaborate or have questions, please contact me via
contact.hannes@nickisch.org or
![]() .
.
Code and Data
gpml
GP The Gaussian Process for Machine Learning toolbox.
 A library for Gaussian process regression and classification for Octave/Matlab
containing a variety of approximate inference schemes ranging from Laplace's
method over expectation propagation to variational Bayes. We furthermore
support large scale approximate inference via the FITC approximation and
MCMC sampling.
A library for Gaussian process regression and classification for Octave/Matlab
containing a variety of approximate inference schemes ranging from Laplace's
method over expectation propagation to variational Bayes. We furthermore
support large scale approximate inference via the FITC approximation and
MCMC sampling.
See the mloss.org project (rank #19),
the gitlab repository,
the JMLR
paper and the gpml
page.
cov = {@covSEiso}; sf = 1; ell = 0.4; hyp.cov = log([ell;sf]);
lik = 'likLaplace'; sn = 0.2; hyp.lik = log(sn);
mean = @meanZero; % set up GP (cov,lik,mean)
nlZ = gp(hyp, 'infEP', mean, cov, lik, X, y); % inference
AwA - Animals with Attributes
CV A dataset for Attribute-Based Classification.
 A dataset for benchmarking transfer-learning or zero-shot learning algorithms. The dataset contains 30475 images in 50 animal classes aligned with Osherson's well-known class/attribute matrix with 85 numerical attributes per class.
A dataset for benchmarking transfer-learning or zero-shot learning algorithms. The dataset contains 30475 images in 50 animal classes aligned with Osherson's well-known class/attribute matrix with 85 numerical attributes per class.
See the download website,
the download website of the free successor dataset,
the CVPR
paper and the IEEE-TPAMI pdf.
OATH - Oslo Aurora THEMIS dateset
CV A dataset for aurora classification.
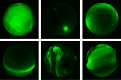 A dataset containing manually labeled aurora images from the THEMIS all-sky imagers. The dataset contains a total of 5824 images in 6 different classes "arc", "diffuse", "discrete", "cloudy", "moon" and "clear". It can be used to train classifiers and benchmark algorithms.
A dataset containing manually labeled aurora images from the THEMIS all-sky imagers. The dataset contains a total of 5824 images in 6 different classes "arc", "diffuse", "discrete", "cloudy", "moon" and "clear". It can be used to train classifiers and benchmark algorithms.
See the download website and
the NIRD archive for the images and a baseline and
the related JGR paper.
glm-ie
ML The Generalised Linear Models Inference and Estimation Toolbox.
 A library for large scale matrix vector multiplication (MVM) based computations
in generalised linear models for Octave/Matlab. We support variational
Bayes, factorial mean field and expectation propagation as well as MAP
estimation using a wide range of penalised least squares solvers for sparse
estimation. A dedicated matrix class provides computational primitives
and a wide range of regularisers are supported.
A library for large scale matrix vector multiplication (MVM) based computations
in generalised linear models for Octave/Matlab. We support variational
Bayes, factorial mean field and expectation propagation as well as MAP
estimation using a wide range of penalised least squares solvers for sparse
estimation. A dedicated matrix class provides computational primitives
and a wide range of regularisers are supported.
See the mloss.org project (rank #35),
the JMLR
paper and the glm-ie
page.
X = matConv2(f, su, 'circ'); % convolution matrix
B = matFD2(su, 'circ'); % finite difference matrix
pot = @potLaplace; s2 = 1e-4; tau = 15;
[m,ga,b,z,nlZ] = dli(X,y,s2,B,pot,tau,opts); % inference
pen = @(s) penAbs(s); % l1-penalty
[u,phi] = plsTN(u0,X,y,B,opt,s2,pen); % estimation
gmm
GP Gaussian mixture modeling with Gaussian process latent variable models and others.
 The toolbox contains code for density estimation using mixtures of Gaussians. Starting from
simple kernel density estimation using spherical and diagonal Gaussian kernels over
manifold Parzen window until mixtures of penalised full Gaussians with only a few
components, the toolbox covers many Gaussian mixture model parametrisation from the recent literature.
Most prominently, the package contains code to use the Gaussian process latent variable model for density estimation.
The toolbox contains code for density estimation using mixtures of Gaussians. Starting from
simple kernel density estimation using spherical and diagonal Gaussian kernels over
manifold Parzen window until mixtures of penalised full Gaussians with only a few
components, the toolbox covers many Gaussian mixture model parametrisation from the recent literature.
Most prominently, the package contains code to use the Gaussian process latent variable model for density estimation.
See the mloss.org project and the corresponding
DAGM paper or get the code
here.
[lp,lpte] = dkde(z,zte); % diagonal kernel density estimation
[lp,lpte] = mpar(z,zte,d,k); % manifold Parzen windows
[lp,lpte] = pgau(z,zte); % penalised Gaussian
fwtn
The fast wavelet transformation for tensor data.
 The code contains a standalone light-weight implementation of the orthonormal
wavelet transform using quadrature mirror filters in C including a Matlab/MEX wrapper.
We fully support D-dimensional
data in L levels. The algorithm has a computational complexity linear in the size
of the input.
The code contains a standalone light-weight implementation of the orthonormal
wavelet transform using quadrature mirror filters in C including a Matlab/MEX wrapper.
We fully support D-dimensional
data in L levels. The algorithm has a computational complexity linear in the size
of the input.
See the mloss.org project
or get the code here.
qmf = [1,1]/sqrt(2); % Haar wavelet
L = 3; % # levels in the pyramid
W = fwtn(X,L,qmf); % apply FWT, inverse: X = fwtn(W,L,qmf,1);
approxXX
GP A variety of approximate inference methods for Gaussian process prediction.
 The code comprises expectation propagation, Laplace's method, the informative vector machine, Gaussian variational mean field,
factorial mean field, on-line expectation propagation, TAP and variational bounding. Note that is
numerically much less robust than the code in gpml. The implementations are meant to illustrate the algorithms
as such and not for use as a black box system in an applied setting.
The functions use the gpml
v2.0 Octave/Matlab interface.
The code comprises expectation propagation, Laplace's method, the informative vector machine, Gaussian variational mean field,
factorial mean field, on-line expectation propagation, TAP and variational bounding. Note that is
numerically much less robust than the code in gpml. The implementations are meant to illustrate the algorithms
as such and not for use as a black box system in an applied setting.
The functions use the gpml
v2.0 Octave/Matlab interface.
See the corresponding
JMLR
paper or get the code here.
hyp = [1; 1]; % ell,sig - GP parameters
cov = {'covSEiso'}; % covariance function
lik = 'cumGauss'; % logistic or cumGauss likelihood
apx = 'LA'; % EP,FV,IVM,KL,LA,LR,OLEP,SO,TAP,TAPnaive or VB
p = binaryGP(hyp, ['approx',apx], cov, lik, x, y, xt); % prediction
Recent Work
Med NN
Dose robustness of deep learning models for anatomic segmentation of computed tomography images
[pdf]
[link]
A. Tsanda, H. Nickisch, T. Wissel, T. Klinder, T. Knopp, M. Grass, JMI, 2024.
Med NN
Learning CT Segmentation from Label Masks Only
[pdf]
[link]
[poster]
A. Tsanda, H. Nickisch, T. Wissel, T. Klinder, T. Knopp, M. Grass, MIDL short paper, 2024.
Med NN
On TotalSegmentator’s performance on low-dose CT images
[pdf]
[slides]
[link]
A. Tsanda, H. Nickisch, T. Wissel, T. Klinder, T. Knopp, M. Grass, SPIE (oral), 2024.
NN
Localized Magnetic Substorm Forecasting using Machine Learning
[pdf]
P. Sado, L. Clausen, W. Miloch, H. Nickisch, 2023.
NN
Substorm Onset Prediction using Machine Learning Classified Auroral Images
[preprint]
[link]
[code]
P. Sado, L. Clausen, W. Miloch, H. Nickisch, Journal of Geophysical Research - Space Weather, 2023.
Scientific Papers
Journal Articles
Med NN
Dose robustness of deep learning models for anatomic segmentation of computed tomography images
[pdf]
[link]
A. Tsanda, H. Nickisch, T. Wissel, T. Klinder, T. Knopp, M. Grass, JMI, 2024.
23) NN
Substorm Onset Prediction using Machine Learning Classified Auroral Images
[preprint]
[link]
[code]
P. Sado, L. Clausen, W. Miloch, H. Nickisch, Journal of Geophysical Research - Space Weather, 2023.
22) Med NN
Cascaded learning in intravascular ultrasound: coronary stent delineation in manual pullbacks
[pdf]
[link]
T. Wissel, K. Riedl, K. Schaefers, H. Nickisch, F. Brunner, N. Schnellbächer, S. Blankenberg, M. Seiffert, M. Grass, Journal of Medical Imaging, 2022.
21) NN
Transfer Learning Aurora Image Classification and Magnetic Disturbance Evaluation
[preprint]
[link]
[code]
P. Sado, L. Clausen, W. Miloch, H. Nickisch, Journal of Geophysical Research - Space Physics, 2021.
20) Med NN
Smart Chest X-ray Worklist Prioritization using Artificial Intelligence: A Clinical Workflow Simulation
[pdf]
[pdf]
[link]
I. Baltruschat, L. Steinmeister, H. Nickisch, A. Saalbach, M. Grass, G. Adam, T. Knopp, H. Ittrich, European Radiology, 2020.
19) Med NN
Learning Metal Artifact Reduction in Cardiac CT Images with Moving Pacemakers Medical Image Analysis
[pdf]
[link]
T. Lossau (Elss), H. Nickisch, T. Wissel, M. Morlock, M. Grass, Medical Image Analysis, 61:101655, 2020.
18) Med NN
Motion Estimation and Correction in Cardiac CT Angiography Images using Convolutional Neural Networks
[pdf]
[link]
T. Lossau (Elss), H. Nickisch, T. Wissel, R. Bippus, H. Schmitt, M. Morlock, M. Grass, Computerized Medical Imaging and Graphics, 76:101640, 2019.
17) GP
Change Surfaces for Expressive Multidimensional Changepoints and Counterfactual Prediction
[pdf]
[link]
W. Herlands, D. B. Neill, H. Nickisch, A. G. Wilson, Journal of Machine Learning Research, 20(99):1−51, 2019.
16) Med NN
Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification
[pdf]
[link]
I. Baltruschat, H. Nickisch, M. Grass, T. Knopp, A. Saalbach, NSR, 9:6381, 2019.
15) Med NN
Motion Artifact Recognition and Quantification in Coronary CT Angiography using Convolutional Neural Networks
[pdf]
[link]
T. Lossau (Elss), H. Nickisch, T. Wissel, R. Bippus, M. Morlock, M. Grass, Medical Image Analysis, 52:68-79, 2019.
14) NN
Automatic classifcation of auroral images from the Oslo Auroral THEMIS (OATH) dataset using machine learning
[pdf]
[link]
[data]
L. Clausen, H. Nickisch, Journal of Geophysical Research - Space Physics, 123(7):5640-5647, 2018.
13) Med
A Functionally Personalized Boundary Condition Model to Improve estimates of fractional flow reserve with CT (CT-FFR)
[pdf]
[link]
M. Freiman, H. Nickisch, H. Schmitt, P. Maurovich-Horvat, P. Donnelly, M. Vembar and L. Goshen, Medical Physics, 45(3):1170-1177, 2018.
12) Med
Improving CCTA based lesions’ hemodynamic significance assessment by accounting for partial volume modeling in automatic coronary lumen segmentation
[pdf]
M. Freiman, H. Nickisch, S. Prevrhal, H. Schmitt, M. Vembar, P. Maurovich-Horvat, P. Donnelly and L. Goshen, Medical Physics, 44(3):1040-1049, 2017.
11) Med
Blind Multi-Rigid Retrospective Motion Correction of MR Images
[pdf]
[link]
A. Loktyushin, H. Nickisch, R. Pohmann and B. Schölkopf, Magnetic Resonance
in Medicine, 73(4):1457-1468, 2015.
10) CV
Attribute-Based Classification for Zero Shot Visual Object Categorization
[pdf]
[link]
C. Lampert, H. Nickisch and S. Harmeling, IEEE Transactions on Pattern
Analysis and Machine Intelligence, 36(3):453-465, 2014.
9) Med Bio
Generating Anatomical Models of the Heart and the Aorta from Medical Images for Personalized Physiological Simulations
[pdf]
[link]
J. Weese, A. Groth, H. Nickisch, H. Barschdorf, F.M. Weber, J. Velut, M. Castro, C. Toumoulin,
J.L. Coatrieux, M. De Craene, G. Piella, C. Tobón-Gomez, A.F. Frangi, D.C. Barber, I. Valverde,
Y. Shi, C. Staicu, A. Brown, P. Beerbaum and D.R. Hose,
Medical and Biological Engineering and Computing, 51(11):1209-1219, 2013.
8) Med
Blind Retrospective Motion Correction of MR Images
[pdf]
[link]
A. Loktyushin, H. Nickisch, R. Pohmann and B. Schölkopf, Magnetic Resonance in Medicine, 70(6):1608-1618, 2013.
7) CV
User-centric Learning and Evaluation of Interactive Segmentation
Systems [pdf]
[link]
P. Kohli, H. Nickisch, C. Rother and C. Rhemann, International Journal
of Computer Vision, 100(3):261-274, 2012.
6) ML
Generating Feature Spaces for Linear Algorithms with Regularized
Sparse Kernel Slow Feature Analysis
[pdf]
[link]
W. Böhmer, S. Grünewälder, H. Nickisch, K. Obermayer, Machine Learning, 89(1):67-86,
2012.
5) ML
glm-ie: The Generalised Linear Models Inference and Estimation
Toolbox [pdf]
[web]
[link]
H. Nickisch, Journal of Machine Learning Research, 13:1699-1703, 2012.
4) ML
Large Scale Bayesian Inference and Experimental Design for Sparse
Linear Models
[pdf]
[link]
M. W. Seeger and H. Nickisch, SIAM Journal on Imaging Sciences, 4(1):166-199,
2011.
3) GP
Gaussian Processes for Machine Learning (GPML) Toolbox
[pdf]
[web]
[link]
C. E. Rasmussen and H. Nickisch, Journal of Machine Learning Research,
11:3011-3015, 2010.
2) Med
Optimization of k-Space Trajectories for Compressed Sensing
by Bayesian Experimental Design
[pdf]
[link]
M. W. Seeger, H. Nickisch, R. Pohmann and B. Schölkopf, Magnetic Resonance
in Medicine, 63(1):116-126, 2010.
1) GP
Approximations for Binary Gaussian Process Classification
[pdf]
[link]
H. Nickisch and C. E. Rasmussen, Journal of Machine Learning Research, 9:2035-2078, 2008.
Peer-Reviewed Conference Papers
32) Med NN
Learning CT Segmentation from Label Masks Only
[pdf]
[link]
[poster]
A. Tsanda, H. Nickisch, T. Wissel, T. Klinder, T. Knopp, M. Grass, MIDL short paper, 2024.
31) Med NN
On TotalSegmentator’s performance on low-dose CT images
[pdf]
[slides]
[link]
A. Tsanda, H. Nickisch, T. Wissel, T. Klinder, T. Knopp, M. Grass, SPIE (oral), 2024, accepted.
30) Med NN
Pulmonary Embolus Detection with Dual-Energy CT Data Augmentation
[pdf]
[slides]
[poster]
C. Hofsäß, R. Gertz, T. Lossau, J. Zemke, T. Knopp, T. Klinder, A. Bunck, H. Nickisch, MIDL short paper (oral), 2022.
29) Med NN
Multi-Resolution 3D convolutional neural networks for automatic coronary centerline extraction in cardiac CT angiography scans
[pdf]
[pdf]
[slides]
Z. Salahuddin, M. Lenga, H. Nickisch, ISBI (oral), 2021.
28) Med NN
Machine-learning based clinical plaque detection using a synthetic plaque lesion model for coronary CTA
[pdf]
[poster]
[slides]
[link]
N. Schnellbächer, H. Ragab, H. Nickisch, T. Wissel, C. Spink, G. Lund, M. Grass, SPIE, 2021.
27) Med NN
Delineation of coronary stents in intravascular ultrasound pullbacks
[pdf]
[link]
T. Wissel, K. Riedl, K. Schaefers, H. Nickisch, F. Brunner, N. Schnellbächer, S. Blankenberg, M. Seiffert, M. Grass, SPIE (oral), 2021.
26) Med NN
How to learn from unlabeled volume data: Self-Supervised 3D Context Feature Learning
[pdf]
[link]
[code]
M. Blendowski, H. Nickisch, M. Heinrich, MICCAI, 2019.
25) Med NN
Dynamic Pacemaker Artifact Removal (DyPAR) from CT Data using CNNs
[pdf]
[link]
T. Lossau, H. Nickisch, T. Wissel, S. Hakmi, C. Spink, M. Morlock, M. Grass, MIDL (oral), 2019.
24) Med NN
When does Bone Suppression and Lung Field Segmentation Improve Chest X-Ray Disease Classification?
[link]
I. Baltruschat, L. Steinmeister, H. Ittrich, G. Adam, H. Nickisch, A. Saalbach, J. von Berg, M. Grass, T. Knopp, ISBI, 2019.
23) GP
State Space Gaussian Processes with Non-Gaussian Likelihood
[pdf]
[poster]
[slides]
[link]
H. Nickisch, A. Solin, A. Grigorievskiy, ICML, 2018.
22) Med NN
2D Motion Estimation in Coronary CT Angiography Images using Convolutional Neural Networks
[pdf]
[poster]
[link]
T. Elss, H. Nickisch, T. Wissel, R. Bippus, M. Morlock, M. Grass, MIDL, 2018.
21) Med NN
Deep-Learning-Based CT Motion Artifact Recognition in Coronary Arteries
[pdf]
[slides]
[link]
T. Elss, H. Nickisch, T. Wissel, H. Schmitt, M. Vembar, M. Morlock, M. Grass, SPIE (oral), 2018.
20) Med NN
Orientation Regression in Hand Radiographs: A Transfer Learning Approach
[pdf]
[link]
I. Baltruschat, A. Saalbach, M. Heinrich, H. Nickisch, S. Jockel, SPIE (poster award), 2018.
19) Med
Nearest Neighbor 3D Segmentation with Context Features
[pdf]
[slides]
[link]
E. Hristova, H. Schulz, T. Brosch, M. Heinrich, H. Nickisch, SPIE (oral), 2018.
18) GP
Scalable Log Determinants for Gaussian Process Kernel Learning
[pdf]
[poster]
[code]
[link]
K. Dong, D. Eriksson, H. Nickisch, D. Bindel, A. G. Wilson, NIPS, 2017.
17) Med
Learning a sparse database for patch-based medical image segmentation
[pdf]
M. Freiman, H. Nickisch, H. Schmitt, P. Maurovich-Horvat, P. Donnelly, M. Vembar, L. Goshen, MICCAI PatchMI, 2017.
16) GP
Scalable Gaussian Processes for Characterizing Multidimensional Change Surfaces
[pdf]
[poster]
[slides]
[link]
W. Herlands, A. G. Wilson, H. Nickisch, S. Flaxman, D. B. Neill, W. van Panhuis, E. Xing, AISTATS, 2016.
15) Med
Automatic coronary lumen segmentation with partial volume modeling improves hemodynamic significance assessment
[pdf]
[slides]
[link]
M. Freiman, Y. Lamash, G. Gilboa, H. Nickisch, S. Prevrhal, H. Schmitt, M. Vembar and L. Goshen, SPIE (oral), 2016.
14) Med ML
SVM-based failure detection of GHT localizations
[pdf]
[slides]
[link]
T. Blaffert, C. Lorenz, H. Nickisch, J. Peters and J. Weese, SPIE (oral), 2016.
13) Med Bio
Learning Patient-Specific Lumped Models for Interactive Coronary Blood Flow Simulations
[pdf]
[talk]
[slides]
H. Nickisch, Y. Lamash, S. Prevrhal, M. Freiman, M. Vembar, L. Goshen and H. Schmitt, MICCAI (oral), 2015.
12) GP
Kernel Interpolation for Scalable Structured Gaussian Processes (KISS-GP)
[pdf,
[supp]
[slides]
[link]
A. G. Wilson and H. Nickisch, ICML, 2015.
11) GP
Fast Kronecker Inference in Gaussian Processes with non-Gaussian Likelihoods
[pdf]
[link]
S. Flaxman, A. G. Wilson, D. B. Neill, H. Nickisch, and A. Smola, ICML, 2015.
10) Med
From Image to Personalized Cardiac Simulation: Encoding Anatomical Structures into a Model-Based Segmentation Framework
[pdf]
[link]
[poster]
H. Nickisch, H. Barschdorf, F. M. Weber, M. W. Krueger, O. Dössel and J. Weese, MICCAI STACOM, 2012.
9) GP
Additive Gaussian Processes
[pdf]
[link]
D. Duvenaud, H. Nickisch and C. E. Rasmussen, NIPS, 2011.
8) ML
Regularized Sparse Kernel Slow Feature Analysis
[pdf]
[link]
W. Böhmer and S. Grünewälder, H. Nickisch and K. Obermayer, ECML/PKDD,
2011.
7) ML
Fast Convergent Algorithms for Expectation Propagation Approximate
Bayesian Inference
[pdf]
[link]
M. W. Seeger and H. Nickisch, AISTATS, 2011.
6) CV
Learning an interactive segmentation system
[pdf]
[link]
H. Nickisch, C. Rother, C. Rhemann and Pushmeet Kohli, ICVGIP, 2010.
Best paper award.
5) GP
Gaussian Mixture Modeling with Gaussian Process Latent Variable
Models
[pdf]
[link]
H. Nickisch and C. E. Rasmussen, DAGM, 2010.
4) ML
Convex variational Bayesian inference for large scale generalized linear models
[pdf]
[link]
H. Nickisch and M. W. Seeger, ICML, 2009.
3) CV
Learning To Detect Unseen Object Classes by Between-Class Attribute
Transfer
[pdf]
[link]
C. H. Lampert, H. Nickisch and S. Harmeling, CVPR, 2009.
2) Med
Bayesian Experimental Design of Magnetic Resonance Imaging Sequences
[pdf]
[link]
M. W. Seeger, H. Nickisch, R. Pohmann and B. Schölkopf, NIPS, 2008.
1) ML
Compressed Sensing and Bayesian Experimental Design
[pdf]
[link]
M. W. Seeger and H. Nickisch, ICML, 2008.
Abstracts and Posters
9) Med NN
Effect of Inter-Observer Variability on Deep Learning in Chest X-Rays
M. Grass, I. M. Baltruschat, A. Saalbach, H. Nickisch, J. von Berg, G. Adam, L. Steinmeister, H. Ittrich, T. Knopp, ECR, C-1093, 2019.
8) Med NN
Effect of Advanced Image Pre-Processing for Multi-Label Chest X-Ray Classification
H. Ittrich, I. M. Baltruschat, L. A. Steinmeister, M. Grass, A. Saalbach, T. Knopp, G. Adam, H. Nickisch, RSNA, AI200-SD-SUA1, 2018.
7) Med NN
Comparison of deep learning-based techniques for organ segmentation in abdominal CT images
[pdf]
[link]
V. Groza, T. Brosch, D. Eschweiler, H. Schulz, S. Renisch, H. Nickisch, MIDL, 2018.
6) GP Med
Dual Compartment Regression for the Generation of Pseudo-CT Images
[pdf]
[link]
A. Schnurr, E. Orasanu, H. Schulz, H. Nickisch, S. Renisch, AAPM, SU-K-601, 2017.
5) Med
Improved On-Site FFR-CT Accuracy by Coronary Tree Standardization [link, p.362]
H. Nickisch, M. Freiman, S. Prevrhal, M. Vembar, P. Donnelly, P. Maurovich-Horvat, L. Goshen, H. Schmitt, ECR, SS 1003, 2017.
4) GP
Is Gun Violence Contagious? Descriptive Spatio-Temporal Testing and Modeling with Large-Scale Point Process
[link]
C. Loeffler, S Flaxman, W. Herlands, H. Nickisch, JSM (Methods for Massive Spatial Data, Statistics and the Environment), 2016.
3) Med NN
Can Pretrained Neural Networks Detect Anatomy? [link]
V. Menkovski, Z. Aleksovski, A. Saalbach, H. Nickisch, NIPS Workshop on Machine Learning in Healthcare, 2015.
2) Med
FID-guided retrospective motion correction based on autofocusing
M. Babayeva, A. Loktyushin, T. Kober, C. Granziera, H. Nickisch, R. Gruetter, G. Krueger, ISMRM-ESMRMB, 2014.
1) ML
Experimental design for genome-wide association studies
[pdf]
C. Lippert, O. Stegle, H. Nickisch, K. Borgwardt, D. Weigel, ISMB, 2010.
Diploma and PhD Theses
2) GP ML
Bayesian Inference and Experimental Design for Large Generalised Linear Models
[pdf]
[link]
H. Nickisch, PhD Thesis, Technische Universität Berlin, Berlin, Germany,
2010.
1) ML
Extraction of visual features from natural video data using Slow Feature Analysis [pdf]
H. Nickisch, Diploma Thesis, Technische Universität Berlin, Berlin, Germany,
2006.
Technical Reports
See arXiv.org for a list of more technical reports.
2) GP
Thoughts on Massively Scalable Gaussian Processes
[link]
A. G. Wilson, C. Dann and H. Nickisch, arXiv.org, 2015.
1) GP
Multiple Kernel Learning: A Unifying Probabilistic Viewpoint
[pdf]
[link]
H. Nickisch and M. W. Seeger, arXiv.org, 2011.
Intellectual Property
See USPTO (inventor = Nickisch AND applicant = Philips) or Justia for the list of my granted US patents and applications (16 grants and 32 applications as of September 2025).
Granted US Patents
16) Resolving and steering decision foci in machine learning-based vascular imaging US11721439B2, 2023-08-08
15) Standardized coronary artery disease metric US11576637B2, 2023-02-14
14) Fractional flow reserve determination US11490867B2, 2022-11-08
13) Coronary artery health state prediction based on a model and imaging data US11301994B2, 2022-04-12
12) Fractional flow reserve simulation parameter customization, calibration and/or training US11195278B2, 2021-12-07
11) Fractional flow reserve determination US11141123B2, 2021-10-12
10) Vascular tree standardization for biophysical simulation and/or an extension simulation for pruned portions US11055845B2, 2021-07-06
9) Apparatus and method for determining a fractional flow reserve US11039804B2, 2021-06-22
8) Mobile FFR simulation US10898267B2, 2021-01-26
7) Heart model guided coronary artery segmentation US10902606B2, 2021-01-26
6) Model-based segmentation of an anatomical structure US10282846B2, 2019-05-07
5) Apparatus for determining a fractional flow reserve value US10258303B2, 2019-04-16
4) Local FFR estimation and visualisation for improved functional stenosis analysis US10111633B2, 2018-10-30
3) Method of determining the blood flow through coronary arteries US9867584B2, 2018-01-16
2) Segmentation apparatus for interactively segmenting blood vessels in angiographic image data US9842401B2, 2017-12-12
1) Processing apparatus for processing cardiac data US9788807B2, 2017-10-17
US Patent Applications
32) FOCUS OPTIMIZATION FOR PREDICTION IN MULTI-FREQUENCY ULTRASOUND IMAGING US20250248693A1, 2025-08-07
31) IMAGE PROCESSING DEVICE, MEDICAL IMAGING SYSTEM AND COMPUTER PROGRAM ELEMENT US20250232449A1, 2025-07-17
30) CONTRAST AGENT ATTENUATION GRADIENT US20250204871A1, 2025-06-26
29) MEASUREMENT OF BLOOD FLOW PARAMETERS US20250160779A1, 2025-05-22
28) A METHOD AND PROCESSOR FOR ANALYZING THE VASCULATURE OF A SUBJECT US20250022133A1, 2025-01-16
27) LOCAL SPECTRAL-COVARIANCE OR LOCAL SPECTRAL COVARIANCE DEFICITS COMPUTATION AND DISPLAY FOR HIGHLIGHTING OF RELEVANT MATERIAL TRANSITIONS IN SPECTRAL CT AND MR US20240090849A1, 2024-03-21
26) METHOD AND SYSTEM FOR VISUALIZATION US20230410307A1, 2023-12-21
25) MEDICAL IMAGE POST-PROCESSING US20230386644A1, 2023-11-30
24) FOCUS OPTIMIZATION FOR PREDICTION IN MULTI-FREQUENCY ULTRASOUND IMAGING US20230148382A1, 2023-05-11
23) UNCERTAINTY-BASED REPRIORITIZATION OF MEDICAL IMAGES BASE UPON CRITICAL FINDINGS US20230077721A1, 2023-03-16
22) FRACTIONAL FLOW RESERVE SIMULATION PARAMETER CUSTOMIZATION, CALIBRATION AND/OR TRAINING US20210110543A1, 2021-04-15
21) METHOD AND APPARATUS FOR PHYSIOLOGICAL FUNCTIONAL PARAMETER DETERMINATION US20210100522A1, 2021-04-08
20) CORONARY ARTERY HEALTH STATE PREDICTION BASED ON A MODEL AND IMAGING DATA US20210073978A1, 2021-03-11
19) SYSTEM AND METHOD FOR IMAGE DECOMPOSITION OF A PROJECTION IMAGE US20200410673A1, 2020-12-31
18) RESOLVING AND STEERING DECISION FOCI IN MACHINE LEARNING-BASED VASCULAR IMAGING US20200411189A1, 2020-12-31
17) INTERACTIVE SELF-IMPROVING ANNOTATION SYSTEM FOR HIGH-RISK PLAQUE BURDEN ASSESSMENT US20200402646A1, 2020-12-24
16) INTRAVASCULAR NAVIGATION USING DATA-DRIVEN ORIENTATION MAPS US20200397294A1, 2020-12-24
15) FRACTIONAL FLOW RESERVE DETERMINATION US20200337664A1, 2020-10-29
14) STANDARDIZED CORONARY ARTERY DISEASE METRIC US20200060637A1, 2020-02-27
13) VASCULAR TREE STANDARDIZATION FOR BIOPHYSICAL SIMULATION AND/OR AN EXTENSION SIMULATION FOR PRUNED PORTIONS US20190318475A1, 2019-10-17
12) APPARATUS AND METHOD FOR DETERMINING A FRACTIONAL FLOW RESERVE US20190209115A1, 2019-07-11
11) FRACTIONAL FLOW RESERVE DETERMINATION US20190083052A1, 2019-03-21
10) MOBILE FFR SIMULATION US20190038356A1, 2019-02-07
9) CT PERFUSION PROTOCOL TARGETING US20180360405A1, 2018-12-20
8) HEART MODEL GUIDED CORONARY ARTERY SEGMENTATION US20180365838A1, 2018-12-20
7) PROCESSING APPARATUS AND METHOD FOR PROCESSING CARDIAC DATA OF A LIVING BEING US20170245824A1, 2017-08-31
6) APPARATUS FOR DETERMINING A FRACTIONAL FLOW RESERVE VALUE US20170105694A1, 2017-04-20
5) MODEL-BASED SEGMENTATION OF AN ANATOMICAL STRUCTURE US20160379372A1, 2016-12-29
4) LOCAL FFR ESTIMATION AND VISUALISATION FOR IMPROVED FUNCTIONAL STENOSIS ANALYSIS US20160302750A1, 2016-10-20
3) PROCESSING APPARATUS FOR PROCESSING CARDIAC DATA US20160206265A1, 2016-07-21
2) SEGMENTATION APPARATUS FOR INTERACTIVELY SEGMENTING BLOOD VESSELS IN ANGIOGRAPHIC IMAGE DATA US20160196660A1, 2016-07-07
1) METHOD OF DETERMINING THE BLOOD FLOW THROUGH CORONARY ARTERIES US20150297161A1, 2015-10-22
Student Supervision and Reviewing
Students (co-)supervised
13) Med NN
Machine Learning in Auroral Image Research; Aurora Image Classification using Machine Learning Techniques and Substorm Forecasting
[disputation]
[pdf]
Pascal Sado, PhD Thesis, University of Oslo, Oslo, Norway, 2023.
12) Med NN
Generalizability of Deep-Learning-Based Pulmonary Embolism Detection from CT Data
[pdf]
Cornelia Hofsäß, Master Thesis, Hamburg University of Technology, Hamburg, Germany, 2022.
11) Med NN
Exploiting Long-Range Dependencies for Prediction of Biomarkers in Coronary Artery Disease
Danil Zamnius, Master Thesis, Universität zu Lübeck, Lübeck, Germany, 2022.
10) Med NN
Deep Learning for Automatic Lung Disease Analysis in Chest X-rays
[pdf]
Ivo Baltruschat, PhD Thesis, Hamburg University of Technology, Hamburg, Germany, 2021.
9) Med NN
Efficient Object Detection in 3D CT Scans
[pdf]
Jannis van Kersbergen, Master Thesis, Eindhoven University of Technology, Eindhoven, The Netherlands, 2021.
8) Med NN
Multi-Resolution 3D Convolutional Neural Networks for Automatic Coronary Centerline Extraction in Cardiac CT Angiography Scans
[link]
[preprint]
[pdf]
Zohaib Salahuddin, Master Thesis, MAIA Erasmus Mundus, Dijon/Cassino/Girona, France/Italy/Spain (best student award), 2020.
7) Med NN
Machine Learning in Cardiac CT Image Reconstruction
[book]
[pdf]
Tanja Lossau, PhD Thesis, Hamburg University of Technology, Hamburg, Germany, 2020.
6) Med NN
Sparse Convolutional Neural Networks in 3D Medical Image Segmentation
Yuan Yao, Master Thesis, Technical University of Berlin, Berlin, Germany, 2019.
5) Med NN
A Tool for Visualizing, Analyzing and Debugging Neural Networks
Laurin Mordhorst, Master Thesis, Hamburg University of Technology, Hamburg, Germany, 2018.
4) Med ML
3D Medical Image Segmentation with Vantage Point Forests and Binary Context Features
[pdf]
Evelin Hristova, Master Thesis, Hamburg University of Applied Sciences, Hamburg, Germany, 2017.
3) Med NN
Deep Learning for Advanced Medical Applications
Ivo Baltruschat, Master Thesis, Universität zu Lübeck, Lübeck, Germany, 2016.
2) Med
Blind Retrospective Motion Correction of MR Images
[link]
[pdf]
[pdf]
Alexander Loktyushin, PhD Thesis, University of Tübingen, Tübingen, Germany, 2015.
1) Med
Model Generation for Automatic Myocardium Segmentation.
Jan Wingerter, Diploma Thesis, Karlsruhe Institute of Technology, Karlsruhe, Germany, 2013.
Reviewing
See
![]() Web of science and
Web of science and
![]() ORCID
for a summary.
ORCID
for a summary.
Machine Learning Conferences
NIPS/NeurIPS (2009-2011, 2013-2024),
ICML (2009, 2011, 2016-2025),
ICLR (2019-2025),
AAAI (2018, 2020),
AISTATS (2009-2012),
COLT (2010),
DAGM/GCPR (2009)
(Medical) Image Processing Conferences
MICCAI (2013, 2018-2025),
MIDL (2019-2020),
ICCV (2009)
Machine Learning Journals
JMLR,
Neurocomputing,
IEEE-TPAMI,
IEEE-TNNLS,
Machine Learning
(Medical) Image Processing Journals
SIIMS,
AIIM,
Medical Image Analysis,
IEEE-TMI,
IEEE-TIP,
IEEE-TBME,
IET-IP,
PMB,
MVAP,
MBEC
Other(s)
Nature Communications,
IEEE-TII,
IEEE-TETC,
IEEE-TMRB,
Physiological Measurement,
CMBBE,
JoVE,
AGU,
JGR Space Physics,
Earth and Space Science